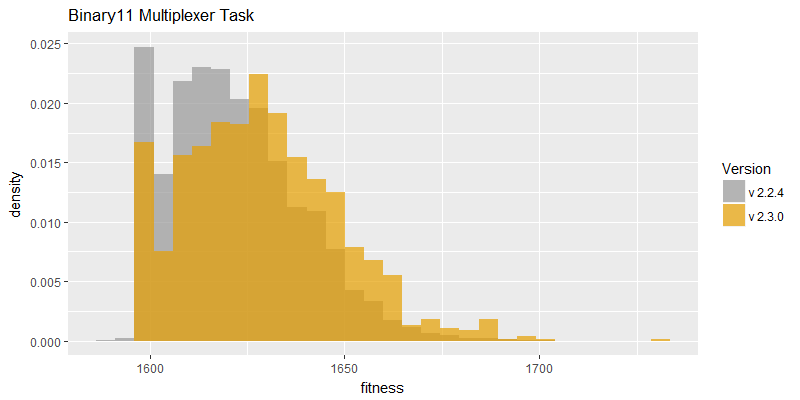
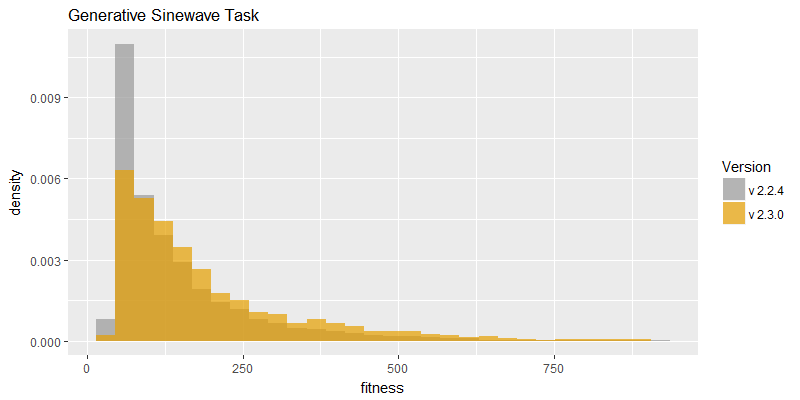
Comparison of best fitness histograms obtained from SharpNEAT v2.2.4 and v2.3.0;
on the Binary11 Multiplexer and Generative Sinewave tasks.
When splitting a connection to add a new node, the connection's innovation ID is looked up in a 'history buffer'. If a connection with the same ID has been split previously then the history buffer holds the IDs of the two new connections and one new node that replaced the connection that was split. These IDs may be re-used for the current connection split, but only if none of the IDs already exist in the genome being operated on; this defect relates to this final test.
The ID lookups are performed using a binary search which returns a negative number if a lookup fails (i.e. there is no such ID in the current genome). However, a test was made for a value of -1 specifically, and not for any negative value. This will have resulted in IDs not being used for a large proportion of instances where they could have been. In effect innovation ID re-use was largely not in effect for the 'add node' mutation.
Lookup of a previously used connection ID between two node IDs (source and target nodes) always failed. A dictionary (hashtable) lookup was used, using the source and target node IDs as the dictionary key; but the key data type was not correctly implemented, resulting in none of the lookups succeeding.
The overall effect of innovation ID re-use is perhaps not well understood, but since SharpNEAT claims to implement this technique it ought to be working correctly until such time as research can be performed to provide evidence for its efficacy (or lack thereof).
A parallelized sub-routine was found to be often in a blocked state (waiting for a thread sync lock). Replacement of that sub-routine with an existing non-parallel equivalent resulted in faster execution overall. Specifically, this relates to the CalculateCentroid() subroutine that is central to the k-means algorithm. Parallelization is still used by calling CalculateCentroid() on each species in parallel, but the sub-routine itself is not parallelized.
In efficacy sampling results were reported for the previous version of SharpNEAT (version 2.2.4). These same tests were re-run against this version (version 2.3.0). The resulting best fitness histograms are provided below. To recap, these histograms show the best fitness achieved on each of a large number of independent SharpNEAT runs, terminating after one minute of execution.


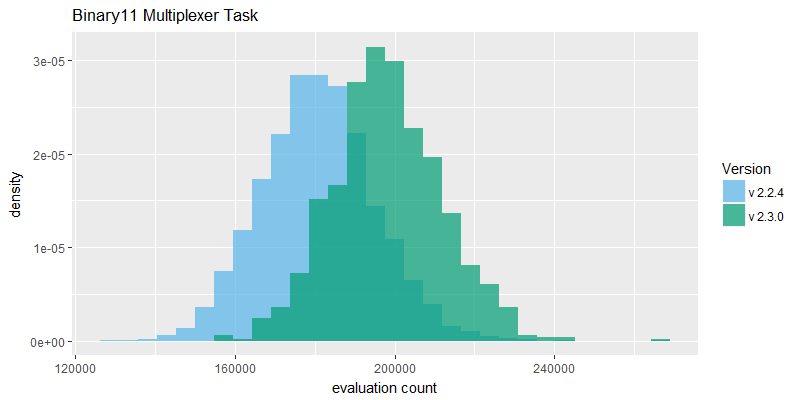
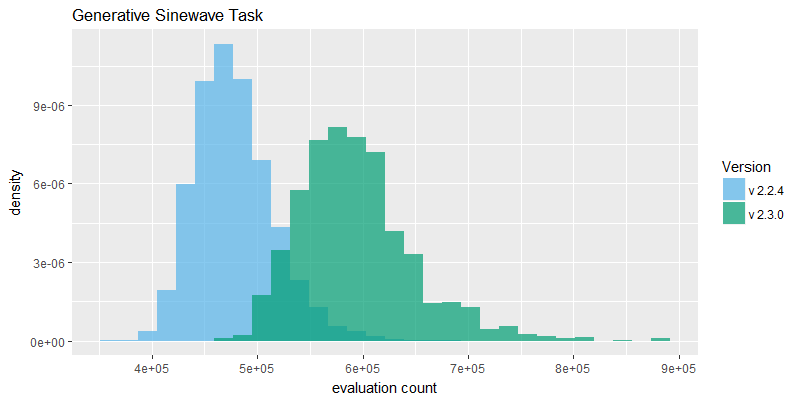
The evaluation count histograms in figure 2 provide good evidence that the overall speed of the NEAT search algorithm has improved in v2.3.0. It is not conclusive evidence because the two fixes may have changed overall size of the evolved genomes such that number of evaluations achieved is not directly correlated with speed of the software. However, the evaluation counts reported are significantly higher on average, on both tasks.
The generative sinewave task in particular exhibits much higher evaluation counts; this is because it derives more benefit from the k-means speciation performance improvement, in turn because proportionally it spends more time in those speciation routines than the binary11 task. The binary11 task is more neural net activation heavy, requiring 2048 activations of a feedforward-only network, per fitness evaluation. In contrast the generative sinewave task requires eighty activations of a cyclic/recurrent neural net. Attaching a performance profiler confirmed this point, with the sinewave task running in v2.2.4 reporting as much as 60% of CPU time blocked on the now improved speciation routine, whereas the binary11 task reported much lower time blocked in that same routine.
The best fitness histograms for both tasks appear to have shifted to the right to some degree, and it seems likely that the speed improvement is the primary cause of this shift, although the defect fixes cannot be ruled out as contributing also. Further efficacy sampling experiments would be required to determine the effect of the fixes, but these were not performed. More work needs to be done to conclusively determine the efficacy of innovation ID re-use.
It is hoped that the efficacy sampling evidence provided demonstrates an empirical improvement in SharpNEAT's search algorithm between v2.2.4 and v2.3.0.
OS Name: Microsoft Windows 10 Home SP0.0 Architecture: 64-bit FrameworkVersion: .NET 4.6.1 (CLR 4.0.30319.42000) CPU Brand: GenuineIntel Name: Intel Core i7-6700T CPU @ 2.80GHz Architecture: x64 Cores: 4 Frequency: 2808 RAM: 16 GB
Colin,
April 29th, 2017
 Copyright 2017 Colin Green.
Copyright 2017 Colin Green.